Repair of Oxidative Damaged DNA
Zharkov, Simmerling and Grollman
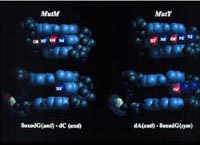 Several DNA repair enzymes in E. coli work in concert to minimize the mutagenic consequences oxidative damage (GO pathway). MutM (Fpg) is a DNA glycosylase/AP lyase that acts preferentially on duplex DNA containing 8-oxodG:dC. MutY is an adenine DNA glycosylase that acts preferentially on 8-oxodG:dA. MutT is an 8-oxodGTPase. Fpg excises 8-oxoguanine from oxidatively-damaged DNA while MutY removes deoxyadenosine paired to 8-oxodG in a DNA replication intermediate, initiating a process by which the oxidized base can later be excised by Fpg. MutT cleanses the cellular deoxynucleoside triphosphate pool, preventing incorporation of 8-oxodGTP into DNA.
Several DNA repair enzymes in E. coli work in concert to minimize the mutagenic consequences oxidative damage (GO pathway). MutM (Fpg) is a DNA glycosylase/AP lyase that acts preferentially on duplex DNA containing 8-oxodG:dC. MutY is an adenine DNA glycosylase that acts preferentially on 8-oxodG:dA. MutT is an 8-oxodGTPase. Fpg excises 8-oxoguanine from oxidatively-damaged DNA while MutY removes deoxyadenosine paired to 8-oxodG in a DNA replication intermediate, initiating a process by which the oxidized base can later be excised by Fpg. MutT cleanses the cellular deoxynucleoside triphosphate pool, preventing incorporation of 8-oxodGTP into DNA.
Bacterial enzymes involved in repair of 8-oxoguanine have functional homologs in eukaryotes. Genes coding for human and mouse homologs of MutT and human MutY have been cloned. Yeast, human and mouse genes coding for an 8-oxoguanine DNA glycosylase (Ogg1) have been isolated by several laboratories. Eukaryotic MutT and MutY proteins are structurally similar to the corresponding E. coli enzymes. However, Ogg1 shows few structural homologies with Fpg.Our research addresses two central questions: (i) how do the several DNA glycosylases involved in repair of 8-oxoguanine in bacteria and mammalian cells recognize their cognate lesions and (ii) what are the biochemical mechanisms by which 8-oxoguanine is efficiently excised from DNA? We analyze the structural features of oxidatively-damaged DNA that confer recognition and binding of 8-oxoguanine by repair enzymes and explore the functional groups and enzymatic mechanisms of the DNA glycosylases involved in these processes.