| Professor Emeritus PhD, Yale University, 1961 158 Life Sciences Building | |
| E-mail: Office: | nicholas.delihas@stonybrook.edu (631) 286-9427 | |
| Publications | ||
Journal editorial membership: Editorial Board Member: International Journal of Molecular Sciences, section 'Molecular Biology' | ||
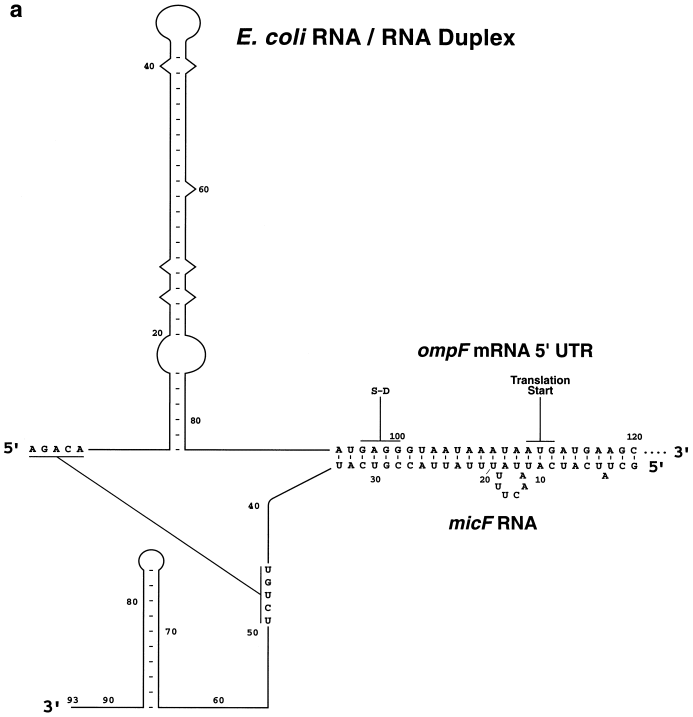
| Past Research | Control of gene expression: Experimental characterization of the first discovered regulatory RNA gene, the micF gene and determination of its transcript functions During past decades, our research concentrated on elucidation of transcriptional activation and suppression of the regulatory non-coding micF RNA gene, characterization of its RNA transcript, and determination of the functional role of the RNA. The micF gene was first discovered and proposed to regulate gene expression by Dr. Masayori Inouye and Dr. Takeshi Mizuno in the early 1980s. Our lab subsequently characterized the micF gene and defined its promoter, found, isolated and characterized the chromosomally encoded RNA transcript, showed in vitro binding of the transcript to the target ompF mRNA, determined the MicF RNA/ target ompF mRNA duplex structure, which showed non-canonical base-pairing (work by Dr. Matthew Schmidt, former graduate student) and showed that the RNA functions as a regulator of gene expression in response to cellular internal and environmental stress conditions. These experimental studies proved that RNA could regulate gene expression. Former graduate student Dr. Janet Andersen played the major role in these determinations. The E. coli MicF RNA has the distinction of being the first non-coding regulatory RNA to have been proposed and characterized. Other labs, led by Dr. Gerhart Wagner in Sweden and Dr. Jorg Vogel in Germany, have shown a greatly expanded role in cell physiology whereby multiple target genes are regulated by micF, including a global regulator that controls ~10% of all protein genes (Mol. Microbiol (2012) 84:414 and 428). In 2024, Stibelman, Sariles and Takahashi determined yet another function of the micF gene in molecular processes, regulation of SeqA, a modulator of DNA replication, and regulation of ObgE, which is important for chromosome partitioning (The Small RNA MicF Represses ObgE and SeqA in Escherichia coli. Microorganisms. 2024;12(12):2397. doi: 10). Also in 2024, Łaska, Matejczyk, and Dauksza developed bacterial biosensors to successfully monitor changes in the environment. They used gene constructs contained the bacterial luciferase gene and the micF gene (The expression of different gene constructs in E. coli SM lux biosensor after exposure to drugs. Sci Rep. 2024 Dec 30;14(1):31899. doi: 10.1038/s41598-024-83190-0.) The initial findings on MicF RNA in the 1980s opened the door to revealing a major principle of molecular genetics: gene expression is regulated by RNA. A write-up of the history of this research appeared in: Nature Methods (October 2022 vol.19 # 10 p1167. How noncoding RNAs began to leave the junkyard. https://www.nature.com/articles/s41592-022-01627-8) First determination of a regulatory non-coding RNA/target mRNA duplex interaction by structure probing determined by Dr. Matthew Schmidt, former graduate student (Schmidt, M., et al., 1995).
| |
| Current Research | Research efforts have now expanded to analysis of human long non-coding RNA (lncRNA) genes and transcripts. lncRNAs are a diverse class of eukaryotic RNAs that have a chain length greater then 200 nucleotides. It is known that the human genome consists of tens of thousands of lncRNA genes and these genes form a major portion of the genome. This offers exciting possibilities for determination of origins and functions, as well as the elucidation of possible roles in disease. We have concentrated on a family of long intergenic non-coding RNA (lincRNA) genes, the FAM230 lncRNA family that are in the critical 22q11.2 region of human chromosome 22. The 22q11.2 region is prone to DNA deletions that result in developmental abnormalities. We have determined that several genes from this RNA gene family are specifically formed in low copy repeats (LCR)s that are found in the 22q11.2 region (see diagram). The LCRs are thought to participate in DNA translocations and non-allelic homologous recombination that lead to 22q11.2 deletions. Although a role for the RNA genes in translocation has not been established, these genes all contain a prominent DNA breakage site and we hypothesize that they may have the potential to participate in DNA translocation. The specificity of these genes also includes expression of the RNA only in a selective tissue, the testes, and this has been established by other labs. In addition to lincRNA gene properties at the DNA level, we have cataloged all the currently known lncRNA genes in the 22q11.2 region (reference The Chemical Biology of Long Noncoding RNAs, 2020, Book chapter) and will attempt to assess lost RNA transcript functions in a chromosomal 22q11.2 deletion with the aim of determining possible roles in abnormal human embryonic development. Chromosomal region 22q11.2. lincRNA genes (#s 1-7) are specifically formed in low copy repeats (LCR)s in the 22q1.2 region and originate from copies of the lincRNA gene FAM230C in LCRs (Delihas, 2018).
The major current research concerns the problem of how protein and lncRNA genes are born. We have detected an ancient element, a nucleation element that forms the basis for the formation several lncRNA genes. With protein genes, the evolutionary progression of open reading frames from ancestral genomes has been analyzed to determine evolutionary root origins of protein sequences that are specific to humans. Found are biased mutations present in non-coding genomic regions where the sequence originated, and biased mutations that are found to extend the sequence to its final length that is present in humans.
| |